Hi all!
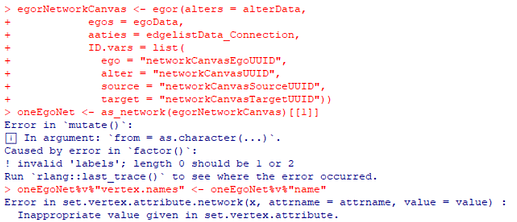
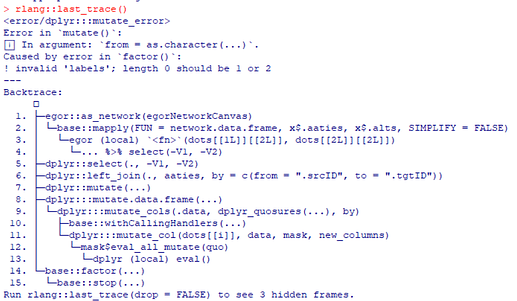
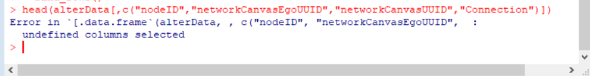
I am working on importing my team’s data into R using this tutorial. I kept running into this error message when I try defining the egor object (screenshot below):
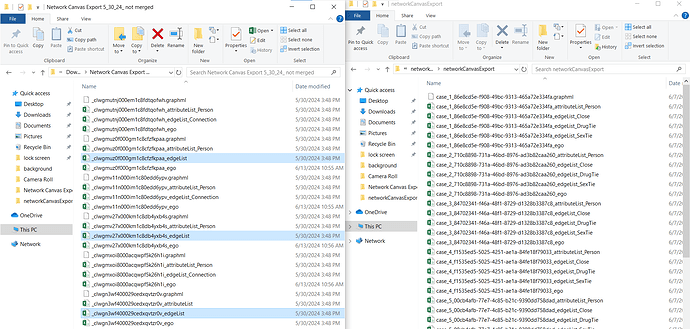
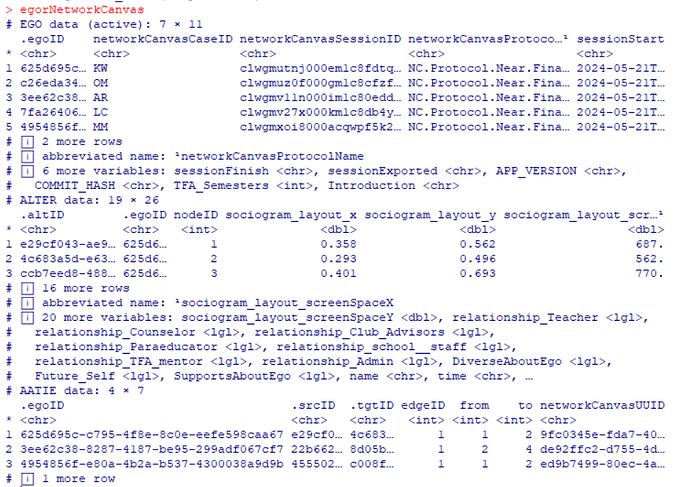
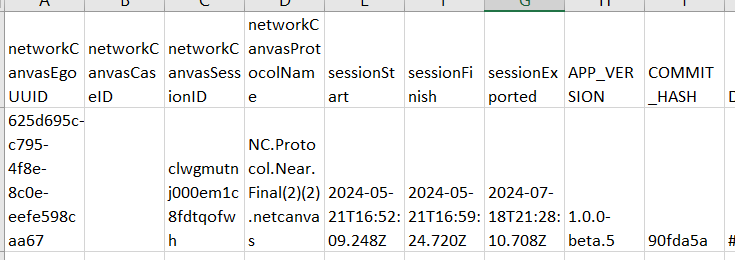
This seemed to suggest that there were issues with those 2 columns, which I was initially confused by since I’d seen those columns were populated properly in the data files named attributeList_Person. So I started comparing our exported data with the demo data, and I found out that we seem to have an extra type of file? (Side-by-side below)
And turns out those extra files only contains the headers mentioned in the error message, but is blank otherwise. So I went ahead and deleted those, in the hopes of resolving the error.
The same error still came up, so I started looking more closely at our exported data. I realized there’s something weird happening with one of the participants’ file clusters – the attributeList_Person is missing completely, and they only have attributeList in the name. Plus, the file is totally blank for some reason. Maybe it’s an exporting error, or the participant didn’t answer all the questions in the protocol?
Either way, I found out which participant it was and I was able to delete their other blank file (edgeList). I tried again, but still ran into that same error. Checking other participants’ files showed that the columns referred to by the error message didn’t yield anything either, as they seem to be properly populated. Removing this participant altogether also didn’t change the error message.
Perhaps I’m barking up the wrong tree with checking the exported files, but the code is identical to the tutorial, save for the segment I replaced when I was cross-comparing the data between my team and the tutorial:
- tutorial:
head(alterData[,c(“nodeID”,“networkCanvasEgoUUID”,“networkCanvasUUID”,“Close”,“Drugs”,“Sex”,“Age”)]) - mine: head(alterData[,c(“nodeID”,“networkCanvasEgoUUID”,“networkCanvasUUID”,“Connection”)])
Any pointers would be greatly appreciated & I’m happy to clarify anything!